I’m currently working with Wikipedia dumps, and to save space, it’s a good thing to make scripts that read directly content from (and write results to) BZipped files.
Setup
Tests where executed on my personal computer:
- i7
- 16GB of ram
On a small Wikipedia dump of 407MB. All timings are in seconds.
bzcat alone
To have a point of comparison, I decompressed the dump using bzcat alone. The timing is 64 seconds.
$ time 1>/dev/null bzcat wikidump.xml.bz2
Using Python
It’s easy enough with Python thanks to the bz2 module that allows to transparently manipulate a compressed file as if it were a normal opened file. Before jumping to Julia, let see how it is done in Python:
from __future__ import print_function
import sys
import bz2
def main():
in_stream = bz2.BZ2File(sys.argv[1])
for line in in_stream:
print(line)
in_stream.close()
if __name__ == "__main__":
main()
Nothing easier, it takes 85 seconds to run.
What about Julia?
Since I really love Julia language, I was tempted to do the same with Julia. Here are the differents solutions that I went through, with their respective timings.
Using bz2 Python module through PyCall
The first naive option is to use the original module from Python. It’s easy enough using the PyCall module. We can install it like so:
julia> Pkg.add("PyCall")
julia> Pkg.update()
The script:
using PyCall
@pyimport bz2
function main()
in_stream = bz2.BZ2File(ARGS[1])
for line in in_stream
println(line)
end
in_stream[:close]()
end
But then we hit the wall… timing is: 1352 seconds. This is likely due to the conversion between Python and Julia datatypes. So not the best option for a data-intensive usage.
Piping result of bzcat to Julia
The second option that came to my mind was: “why not using bzcat?”. It’s easy enough, we just have to read from STDIN:
function main()
for line in eachline(STDIN)
print(line)
end
end
Here is the invocation:
$ bzcat wikidump.xml.bz2 | julia bz2_bench.jl
Timing is now a more reasonable 72 seconds. So that is less than the Python version shown above. But this is not satisfactory enough. Why not use the wonderful capabilities of Julia to run external commands an pipe results?
Invoking bzcat from Julia
It is easy to invoke commands from inside Julia using backquotes: run(`cmd`) and |> to pipe between commands and streams. Let’s do it:
function main()
file = ARGS[1]
run(`bzcat $(file)` |> STDOUT)
end
The script is equivalent to: bzcat wikidump.xml.bz2, but it’s quite impressive to see how easy it is to do this inside a Julia script. This time is about 66 seconds, more or less the same than with the external piping from bzcat.
But it would be useful to get lines of contents from the stream, like it was in the original Python script. For this task, Julia standard library offers a multitudes of handy functions. The one we will use is readsfrom that returns two things: stdout of the given process, and the process itself. Here it is in action:
function main()
file = ARGS[1]
stdout, p = readsfrom(`bzcat $(file)`)
for line in eachline(stdout)
print(line)
end
end
Timing is now about 74 seconds, this is 10 seconds faster than the first Python version. But we don’t rely on a module. Instead, we make use of the ability to play with command invocations, stream pipings, and the like that Julia allows.
Timings
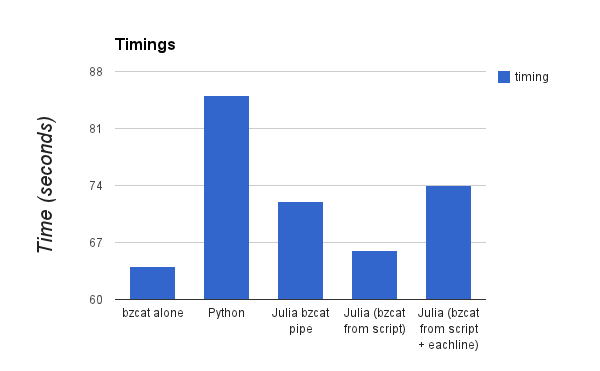
Timings are relatively close since the big work is done in the decompression, that’s why there isn’t much difference between Julia and Python.
Conclusion
I was first tempted to implement a Julia wrapper over bzlib, but what for? When it’s so easy to invoke external commands and manipulate their input and output streams. Julia is a young language, but it’s so flexible and extensible, that often I forget about it!